x <- feature_selection(
ranger::ranger,
data = data,
test = test,
response = "label",
# stat = function(x) quantile(x, .25),
stat = function(x) quantile(x, .75),
iterations = iter,
sample_frac = frac,
predict_function = function(object, newdata){ranger:::predict.ranger(object, data = newdata)$predictions},
parallel = FALSE,
max.depth = 15
)
#> ℹ Using 1 - accuracy as loss function.
#> ℹ Fitting 1st model using 500 predictor variables.
#>
#> ── Round #1 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
#> ℹ Removing 4 variables. Fitting new model with 496 variables.
#>
#> ── Round #2 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
#> ℹ Removing 9 variables. Fitting new model with 487 variables.
#>
#> ── Round #3 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
#> ℹ Removing 108 variables. Fitting new model with 379 variables.
#>
#> ── Round #4 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
#> ℹ Removing 5 variables. Fitting new model with 374 variables.
#>
#> ── Round #5 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
#> ℹ Removing 18 variables. Fitting new model with 356 variables.
#>
#> ── Round #6 ──
#>
#> ℹ Using `dplyr::sample_frac` as sampler.
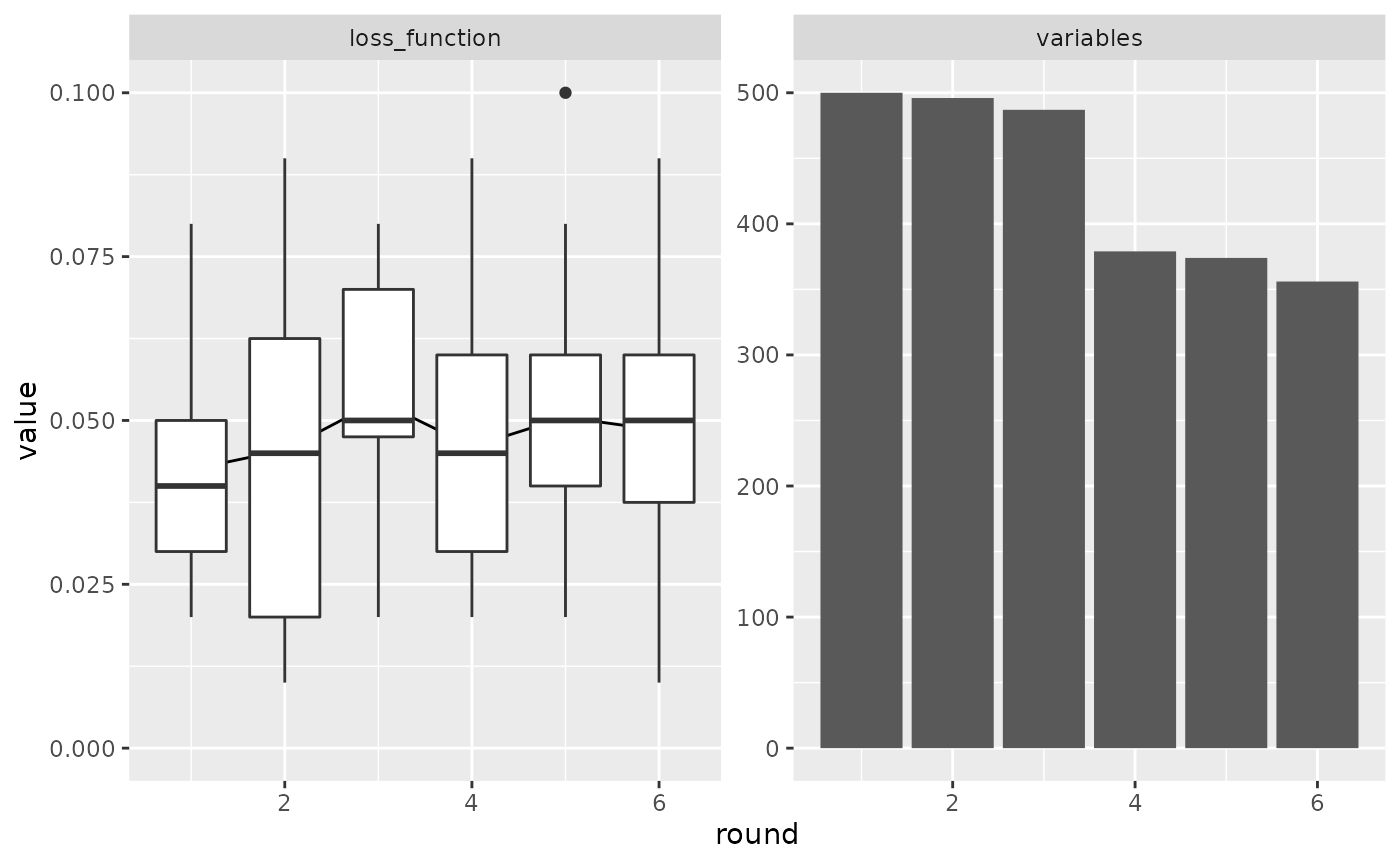
x
#> # A tibble: 6 × 5
#> round mean_value values n_variables variables
#> <dbl> <dbl> <list> <int> <list>
#> 1 1 0.0425 <dbl [20]> 500 <chr [500]>
#> 2 2 0.0455 <dbl [20]> 496 <chr [496]>
#> 3 3 0.053 <dbl [20]> 487 <chr [487]>
#> 4 4 0.046 <dbl [20]> 379 <chr [379]>
#> 5 5 0.0505 <dbl [20]> 374 <chr [374]>
#> 6 6 0.0485 <dbl [20]> 356 <chr [356]>
plot(x)